Anti-cancer drug also functions as COVID-19 antiviral therapy
Johns Hopkins Medicine study suggests small molecule inhibitor RK-33 can block multiple variants of the SARS-CoV-2 virus from replicating.
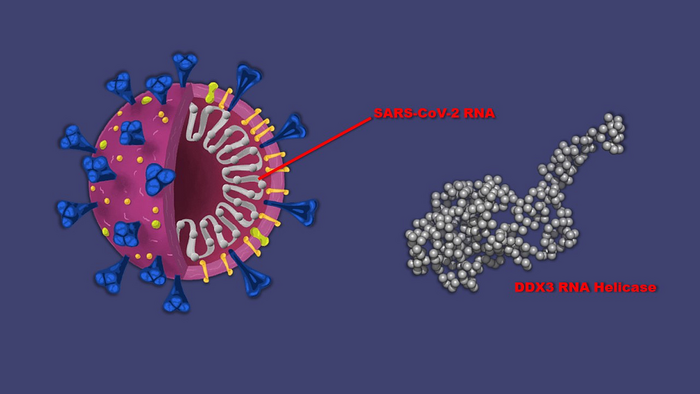
[Sept 22, 2022: Michael Newman, Johns Hopkins Medicine]
Cutaway view (left) of SARS-CoV-2, the virus that causes COVID-19, showing its double stranded RNA genome. During infection, the virus takes over a host cell’s DDX3 RNA helicase protein (right) to unwind the viral RNA and enable it to be copied. A new study shows that a Johns Hopkins Medicine-created protein, RK-33, can prevent DDX3 from carrying out this function. (CREDIT: Graphic created by M.E. Newman, Johns Hopkins Medicine, using SARS-CoV-2 graphic courtesy of Davian Ho for the Innovative Genomics Institute and DDX3 graphic courtesy of the Small Angle Scattering Biological Data Bank. Scale of virus to RK-33 is not proportional)
Based on findings from a new study by a Johns Hopkins Medicine-led research team, an effective means of fighting SARS-CoV-2, the virus that causes COVID-19, may be possible that circumvents the problem of waning immunity often observed when current vaccines deal with emerging COVID variants.
The method uses a small molecule inhibitor (a molecule approximately 1 nanometer in size that inhibits specific interactions between proteins) called RK-33 to block the virus’s ability to take over a host cell’s “genetic manufacturing plant” and make copies of itself.
“To date, COVID-19 vaccines have relied on preventing the binding of a SARS-CoV-2 surface protein — called the spike protein — to host cells and enabling infection, but if the spike protein changes with new variants, a vaccine’s effectiveness may be weakened,” says study senior author Venu Raman, Ph.D., professor of radiology, oncology and pharmacology at the Johns Hopkins University School of Medicine. “In contrast, our study shows that RK-33’s antiviral capability is unaffected by spike protein mutations and remains consistent across four SARS-CoV-2 variants.”
The research was first posted in the journal Frontiers in Microbiology.
Related Stories:
For several years, Raman and his colleagues have studied a protein known as DDX3 and its impact on cancer. DDX3 is a ribonucleic acid (RNA) helicase, a protein that unwinds the double-stranded RNA controlling many tumor cells, enabling the RNA’s genetic code to be read (or translated).
This, in turn, leads to the creation of new cancer cells and malignant spread of the disease. Studies by Raman’s team and others have suggested that RK-33, a DDX3 inhibitor developed in the Raman laboratory, can slow down cancer progression by keeping RNA from unwinding for translation.
DDX3 protein also has been shown to help promote the infectivity of many RNA viruses, such as HIV and respiratory syncytial virus (RSV). Consequently, RK-33, the DDX3-inhibitor with great promise as a cancer fighter, is now being seriously considered for a second therapeutic function: a broad-spectrum antiviral agent.
Comparison of mutation status of SARS-CoV-2 variants. (A) Radar plot displaying estimated protein sequence evolutionary distance to the Lineage A variant for the Alpha (purple), Beta (green), and Delta (blue) variants. Only proteins that contain substitutions in at least one of the variants are shown. Proteins are ordered to group similar distances together for each isolate. *Delta ORF7a protein sequence has an estimated distance of 0.67 from Lineage A due to a deletion resulting in a frameshift. **Delta ORF8 protein sequence has an estimated distance of 0.02 from the Lineage A variant. These distances are plotted at the limits of the plot for clarity. (B) Comparison of the SARS-CoV-2 proteome across the four variants of varying lineage used in this study. Arrows indicate all proteins encoded by the SARS-CoV-2 genome, and vertical lines denote amino acid variations with respect to the Lineage A variant. (CREDIT: journal Frontiers in Microbiology)
“We know that many RNA viruses usurp the DDX3 helicase function of the host cell to facilitate their own replication,” says Raman. “When scientific studies revealed that small concentrations of RK-33 blocked replication and limited infectivity by human parainfluenza type 3 virus, RSV, dengue virus, Zika virus and West Nile virus — and potentially, HIV — our team decided to see whether RK-33 could work on SARS-CoV-2 as well.”
Along with testing RK-33’s impact on SARS-CoV-2 infectivity and reproduction, the researchers extended their study to determine if the inhibitory action observed was limited to specific variants of the virus or would be effective against multiple variants.
Comparison of mutation status of SARS-CoV-2 variants. (C) Multiple alignment of SARS-CoV-2 spike protein sequences for the four variants shown in (B) displayed in greater detail. Amino acid variations with respect to the Lineage A variant are indicated with red vertical lines. (D) Spike protein homotrimer homology model of Lineage A variant with amino acid variations colored red for the Alpha, Beta, and Delta variants. (CREDIT: journal Frontiers in Microbiology)
They used RK-33 to target DDX3 in laboratory cells infected with four variants of SARS-CoV-2 — the original virus and the alpha, beta and delta variants.
“Our results indicate that for the four SARS-CoV-2 variants we tested, RK-33 treatment of infected cells showed significant reductions in the viral load [the number of virus particles in a defined sample size], as much as a thousandfold,” says Raman. “Consistent with this finding, we saw a downregulation [reduction in production] of most SARS-CoV-2 proteins and genes, including the protein transmembrane serine protease 2 [TMPRSS2], which we know strongly participates in the infectivity and spread of coronaviruses.”
Raman adds that not only did RK-33 work with four different SARS-CoV-2 variants, the protein’s antiviral activity is unaffected by the mutations that created each of them.
“Vaccines designed against the spike protein of one SARS-CoV-2 variant may not be as effective if a new variant has a mutated spike protein,” he explains. “The ability of RK-33 to inhibit DDX3’s unwinding of viral RNA for translation is independent of the spike protein, so it should remain effective against most variants.”
Currently, Raman and his team are looking at RK-33 as an antiviral against the omicron variant of SARS-CoV-2. The researchers hope to publish their findings later this year.
Along with Raman, the Johns Hopkins Medicine members of the research team are Farhad Vesuna (joint study lead author), Robert Scharpf and Paul Winnard. Collaborators from the Virginia Polytechnic Institute and State University are Ivan Akhrymuk (joint study lead author), Kylene Kehn-Hall, Lauren Panny and Amy Smith. Shih-Chao Lin from the National Taiwan Ocean University also contributed to the study.
The research was supported by National Institutes of Health grant R01CA207208 and the Flight Attendant Medical Research Institute.
Raman holds a patent on the composition of RK-33. The other study authors report no conflicts of interest.
Note: Materials provided above by Johns Hopkins Medicine. Content may be edited for style and length.
Like these kind of feel good stories? Get the Brighter Side of News' newsletter.



